Input data and parameters
QualiMap command line
| qualimap bamqc -bam E2348-69.bam -c -nw 400 -hm 3 |
Alignment
| Command line: | bwa mem -R @RG\tID:E2348-69\tSM:E2348-69\tLB:E2348-69\tPL:ILLUMINA -t 8 /srv/GT/reference/Escherichia_coli_O127H6_EPEC-E2348_69-V2/Ensembl/EPEC-E2348_69-V2/Sequence/BWAIndex/genome.fa /scratch/BWA_2025-02-06--09-30-17_E2348-69_temp3827628/E2348-69-trimmed_R1.fastq.gz /scratch/BWA_2025-02-06--09-30-17_E2348-69_temp3827628/E2348-69-trimmed_R2.fastq.gz |
| Draw chromosome limits: | yes |
| Analyze overlapping paired-end reads: | no |
| Program: | bwa (0.7.18-r1243-dirty) |
| Analysis date: | Thu Feb 06 09:43:08 CET 2025 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | no |
| Number of windows: | 400 |
| BAM file: | E2348-69.bam |
Summary
Globals
| Reference size | 4,848,621 |
| Number of reads | 5,827,464 |
| Mapped reads | 5,534,444 / 94.97% |
| Unmapped reads | 293,020 / 5.03% |
| Mapped paired reads | 5,534,444 / 94.97% |
| Mapped reads, first in pair | 2,780,723 / 47.72% |
| Mapped reads, second in pair | 2,753,721 / 47.25% |
| Mapped reads, both in pair | 5,500,026 / 94.38% |
| Mapped reads, singletons | 34,418 / 0.59% |
| Secondary alignments | 0 |
| Supplementary alignments | 14,114 / 0.24% |
| Read min/max/mean length | 18 / 151 / 135.24 |
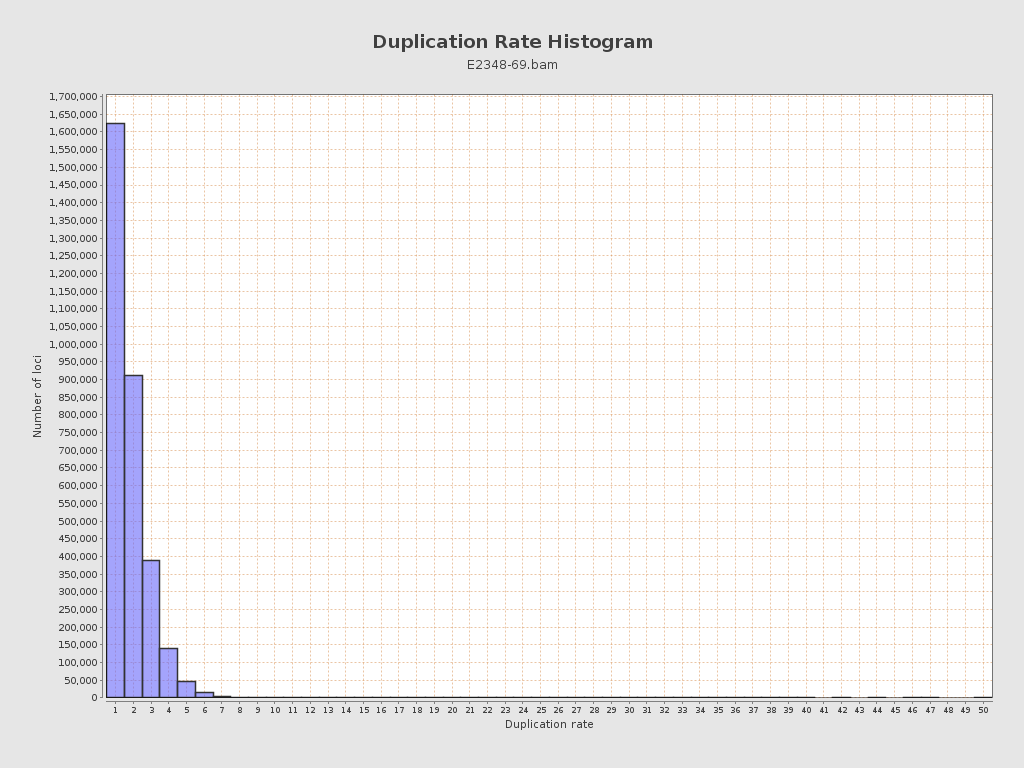
| Duplicated reads (flagged) | 191,153 / 3.28% |
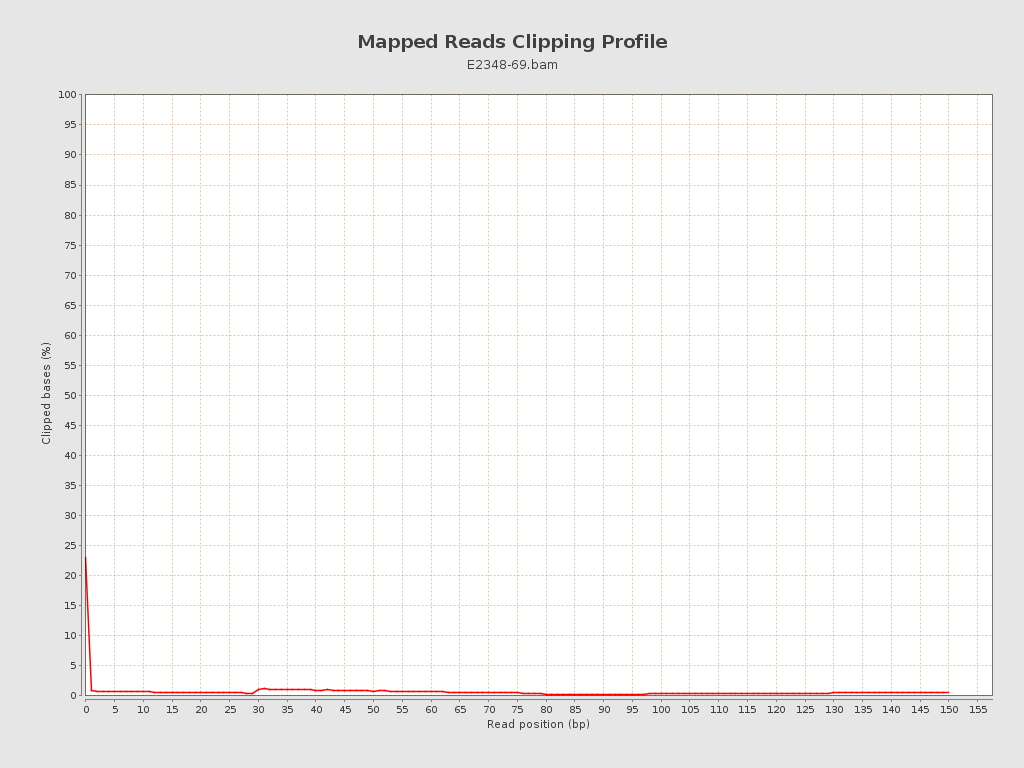
| Clipped reads | 57,003 / 0.98% |
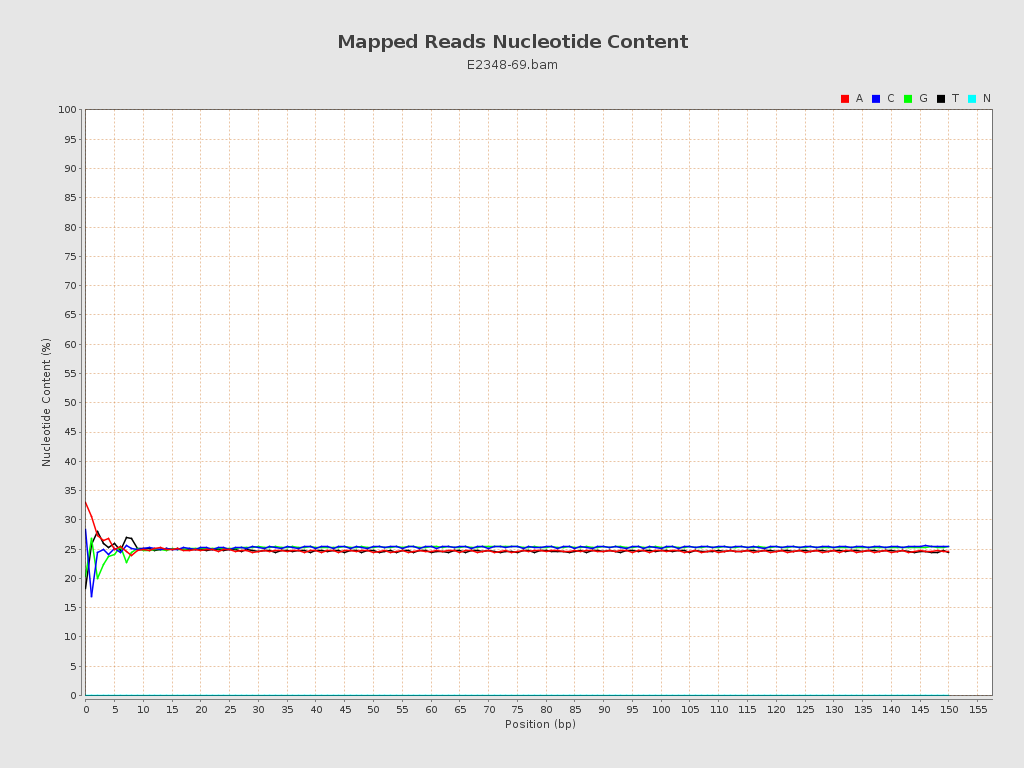
ACGT Content
| Number/percentage of A's | 186,537,405 / 24.84% |
| Number/percentage of C's | 189,585,353 / 25.25% |
| Number/percentage of T's | 185,668,010 / 24.73% |
| Number/percentage of G's | 189,012,413 / 25.17% |
| Number/percentage of N's | 14,158 / 0% |
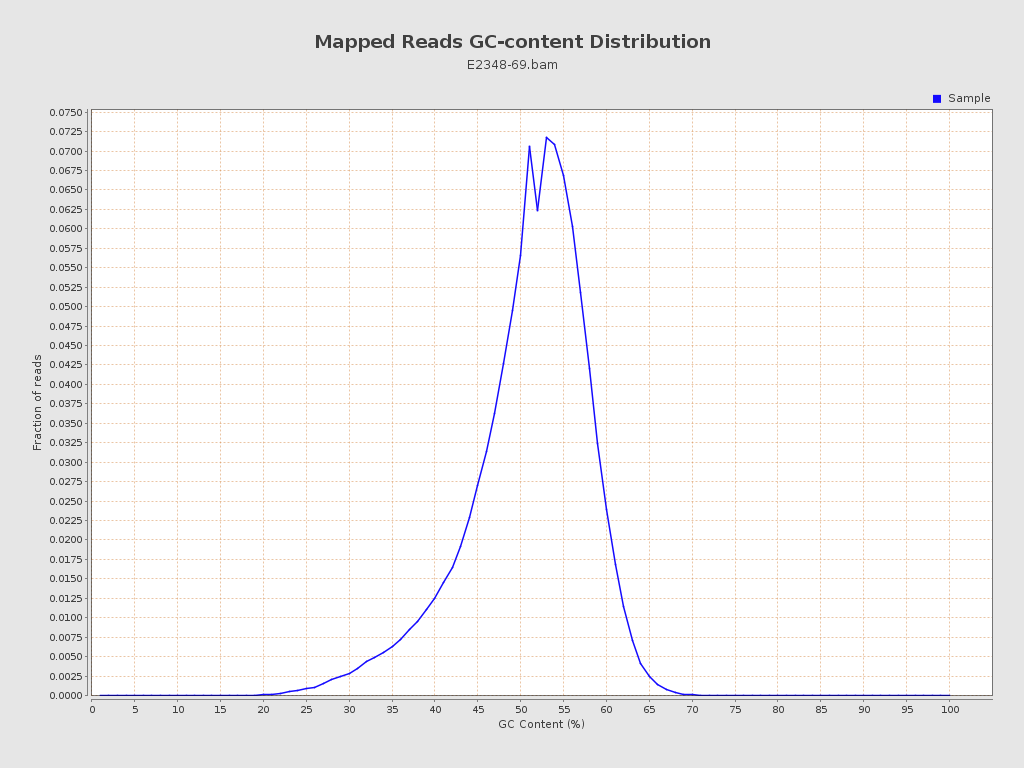
| GC Percentage | 50.42% |
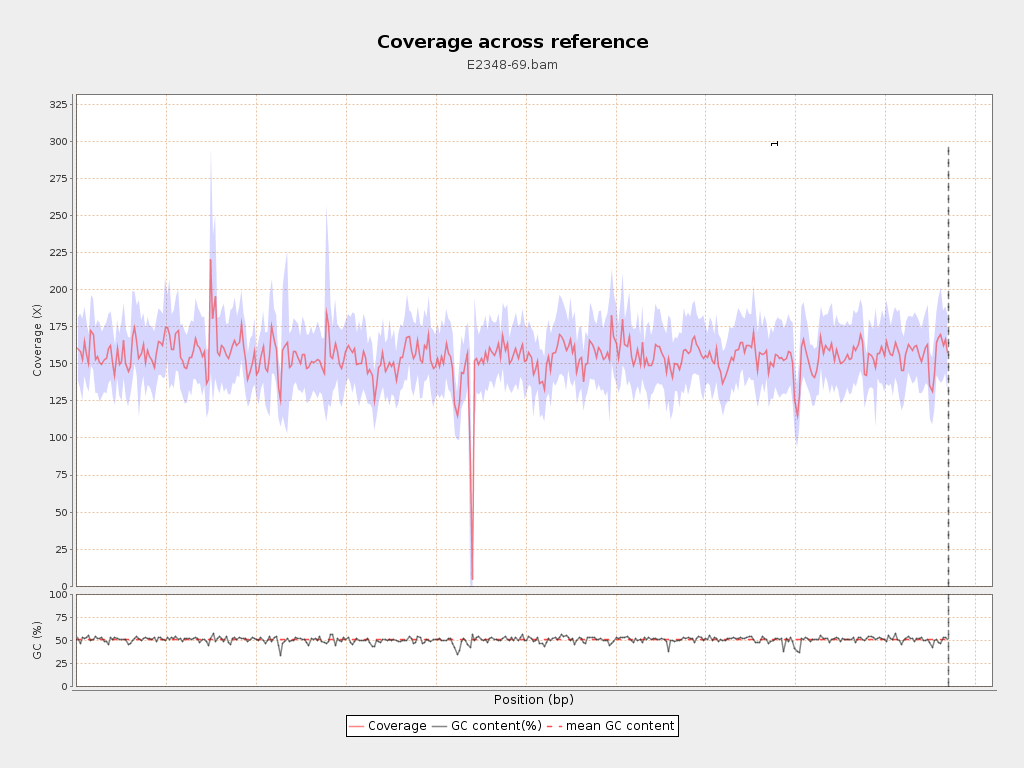
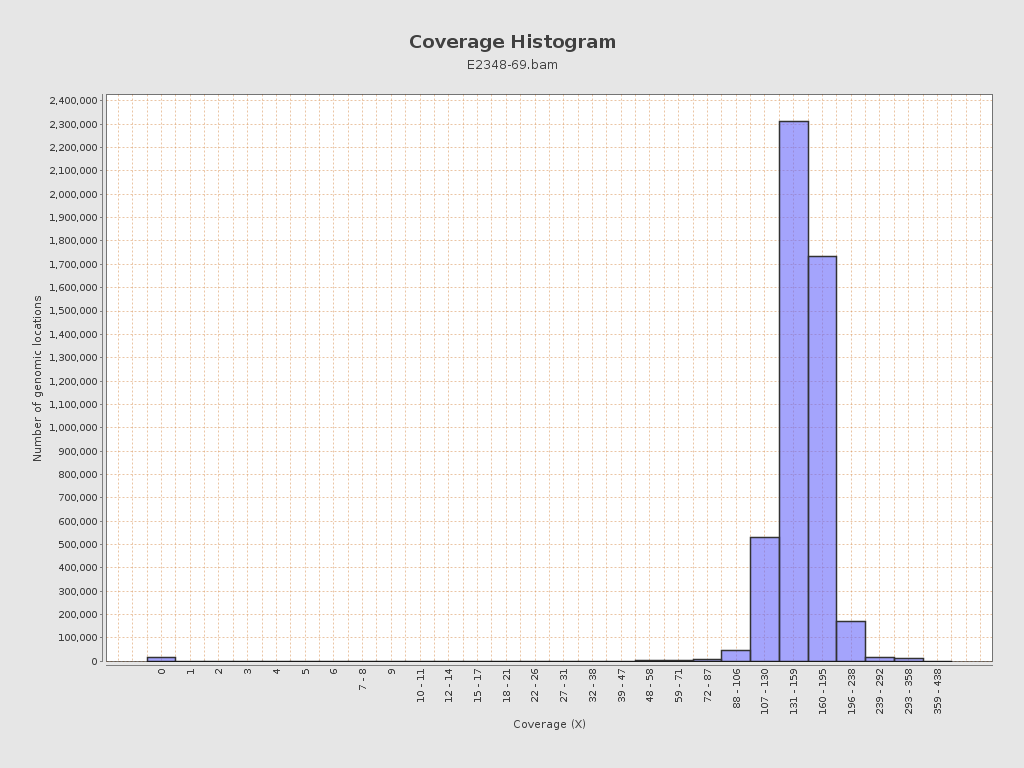
Coverage
| Mean | 154.854 |
| Standard Deviation | 25.6678 |
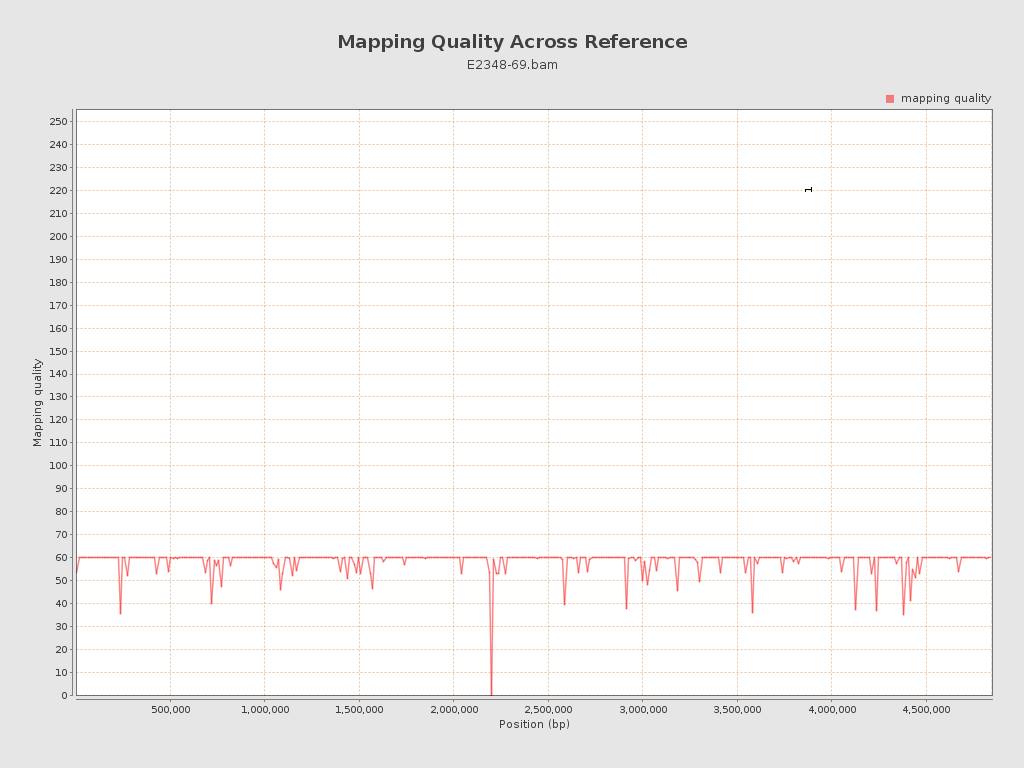
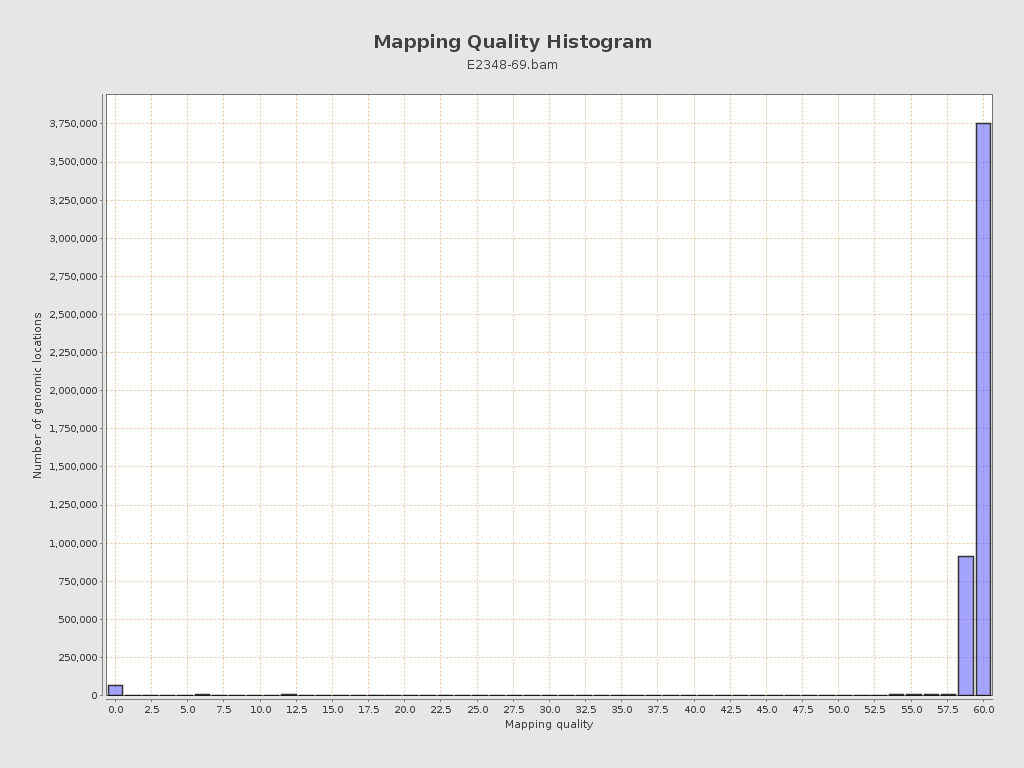
Mapping Quality
| Mean Mapping Quality | 58.49 |
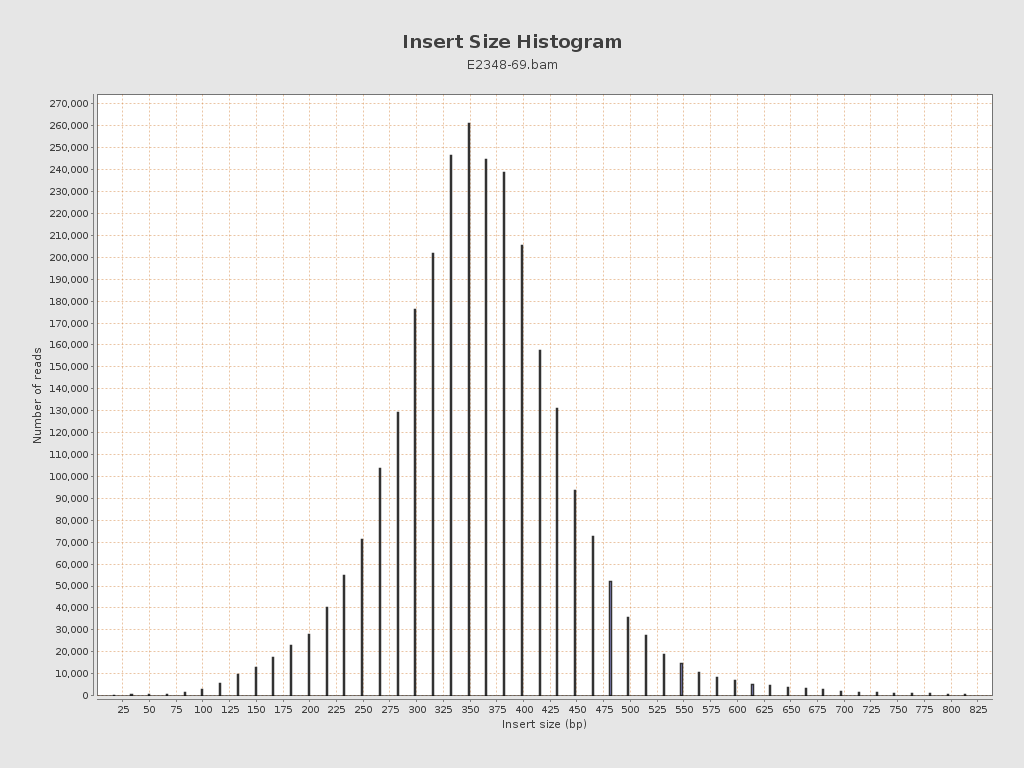
Insert size
| Mean | 13,126.96 |
| Standard Deviation | 176,657.36 |
| P25/Median/P75 | 317 / 365 / 415 |
Mismatches and indels
| General error rate | 0.16% |
| Mismatches | 1,218,318 |
| Insertions | 2,213 |
| Mapped reads with at least one insertion | 0.04% |
| Deletions | 6,690 |
| Mapped reads with at least one deletion | 0.12% |
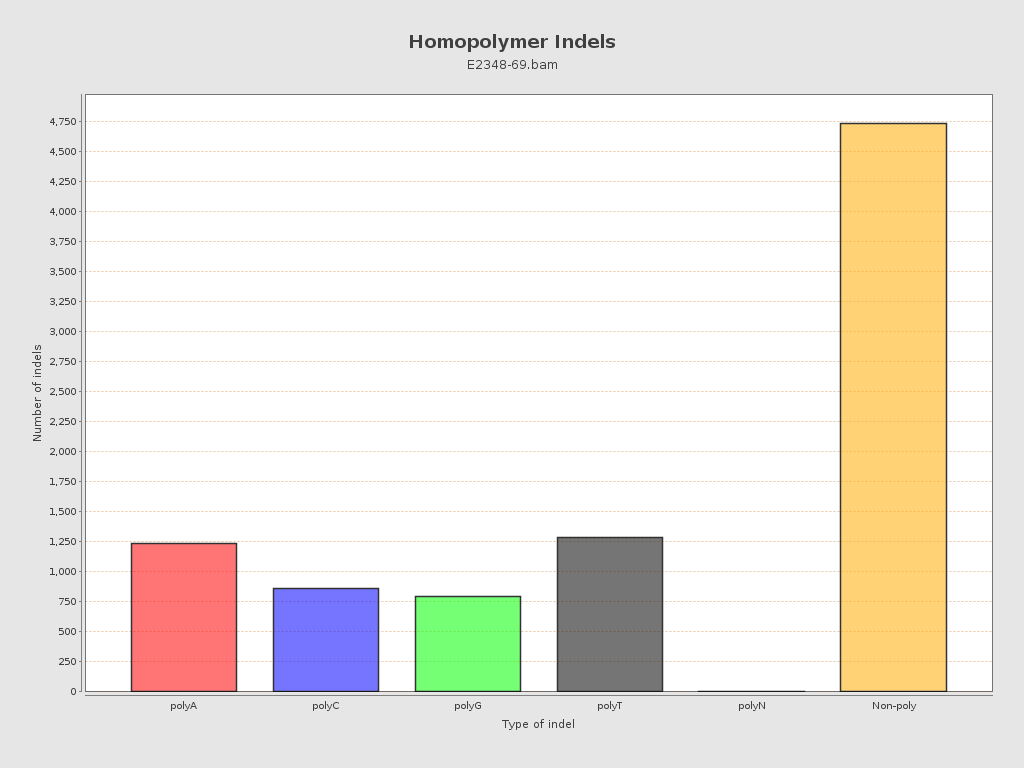
| Homopolymer indels | 46.83% |
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| 1 | 4848621 | 750828167 | 154.854 | 25.6678 |